User’s guide
…or IDT’s list of opinionated howtos
This section seeks to provide users of the Mila infrastructure with practical knowledge, tips and tricks and example commands.
Quick Start
Users first need login access to the cluster. It is recommended to install milatools which will help in the set up of the ssh configuration needed to securely and easily connect to the cluster.
mila code
milatools also makes it easy to run and debug code on the Mila cluster.
First you need to setup your ssh configuration using mila init. The
initialisation of the ssh configuration is explained
here and in the mila init section of github page.
Once that is done, you may run VSCode
on the cluster simply by using the Remote-SSH extension
and selecting mila-cpu as the host (in step 2).
mila-cpu allocates a single CPU and 8 GB of RAM. If you need more
resources from within VSCode (e.g. to run a ML model in a notebook), then
you can use mila code. For example, if you want a GPU, 32G of RAM and 4 cores,
run this command in the terminal:
mila code path/on/cluster --alloc --gres=gpu:1 --mem=32G -c 4
The details of the command can be found in the mila code section of github page. Remember that you need to
first setup your ssh configuration using mila init before the mila code
command can be used.
Logging in to the cluster
To access the Mila Cluster clusters, you will need a Mila account. Please contact Mila systems administrators if you don’t have it already. Our IT support service is available here: https://it-support.mila.quebec/
You will also need to complete and return an IT Onboarding Training to get access to the cluster. Please refer to the Mila Intranet for more informations: https://sites.google.com/mila.quebec/mila-intranet/it-infrastructure/it-onboarding-training
IMPORTANT : Your access to the Cluster is granted based on your status at Mila (for students, your status is the same as your main supervisor’ status), and on the duration of your stay, set during the creation of your account. The following have access to the cluster : Current Students of Core Professors - Core Professors - Staff
SSH (Secure Shell)
All access to the Mila cluster is via SSH using public-key authentication. As of March 31, 2025, this will become the only means of authentication, and password-based authentication will no longer work.
SSH key authentication is a technique using pairs of closely-linked keys: A private key, and a corresponding public key. The public key should be distributed to everyone, while the private key is known to only one person. The public key can be used by anyone to challenge a person to prove their identity. If they have the corresponding private key, that person can perform an electronic signature that everyone can validate but that no one else could have done themselves. The challenge is thus answered by demonstrating possession of the private key (and therefore their identity), without ever revealing the private key itself.
Mila asks you to generate a pair of SSH keys, to provide Mila only with your public key, which has no confidentiality implications, and to keep the private key for yourself. The private key must remain secret and solely known to you, because anyone who possesses it is capable of impersonating you by performing your electronic signature.
During the IT Onboarding Training, you will be asked to submit that SSH public key.
If you do not know what SSH keys are, or are not familiar with them, you can read the informative material below, then proceed to generate them.
If you do not already have SSH keys, or are not sure if you have them, skip to the instructions on how to generate them here.
If you do have SSH keys, you can skip to configuring SSH for access to Mila.
Logging in with SSH
Login to the Mila cluster is with ssh through four Internet-facing
login nodes and a load-balancer. At each connection through the load-balancer,
you will be redirected to the least loaded login node.
# Generic login, will send you to one of the 4 login nodes to spread the load
ssh -p 2222 <user>@login.server.mila.quebec
# To connect to a specific login node, X in [1, 2, 3, 4]
ssh -p 2222 <user>@login-X.login.server.mila.quebec
This is a significant amount of typing. You are strongly encouraged to add
a mila “alias” to your SSH configuration file (see below
for how). With a correctly-configured SSH you can now simply run
# Login with SSH configuration in place
ssh mila
# Can also scp... vvvv
scp file-to-upload.zip mila:scratch/uploaded.zip
# vvvv ... and rsync!
rsync -avz mila:my/remote/sourcecode/ downloaded-source/
to connect to a login node. The mila alias will be available to ssh,
scp, rsync and all other programs that consult the SSH configuration file.
Upon first login, you may be asked to enter your SSH key passphrase. Use the passphrase you used to create your SSH key below.
Upon first login, you may also be asked whether you trust the *Mila* login servers’ own SSH keys. The ECDSA, RSA and ED25519 fingerprints for Mila’s login nodes are:
SHA256:baEGIa311fhnxBWsIZJ/zYhq2WfCttwyHRKzAb8zlp8 (ECDSA)
SHA256:Xr0/JqV/+5DNguPfiN5hb8rSG+nBAcfVCJoSyrR0W0o (RSA)
SHA256:gfXZzaPiaYHcrPqzHvBi6v+BWRS/lXOS/zAjOKeoBJg (ED25519)
If the fingerprints presented to you do not match one of the above, do not trust them!
Tip
You can run commands on the login node with ssh directly, for example
ssh mila squeue -u '$USER' (remember to put single quotes around any
$VARIABLE you want to evaluate on the remote side, otherwise it will be
evaluated locally before ssh is even executed).
Important
Login nodes are merely entry points to the cluster. They give you access to the compute nodes and to the filesystem, but they are not meant to run anything heavy. Do not run compute-heavy programs on these nodes, because in doing so you could bring them down, impeding cluster access for everyone.
This means no training scripts or experiments and no compilation of software unless it is small or ends quickly. Do not run anything that demands a sustained large amount of computation or a large amount of memory.
Rule of thumb: Never run a program that takes more than a few seconds on a login node, unless it mostly sleeps or mostly moves data.
Examples: A non-exhaustive list of use-cases, to give a sense of what is and is not allowed on the login nodes:
A Python training script is unacceptable on the login nodes. (Too computationally- and memory-intensive)
A Python or shell script that downloads a dataset and exits immediately after may be acceptable on the login nodes. (Mostly moves data)
A Python hyperparameter search script that uses
submititto launch jobs and only sleeps waiting for them to end and run other jobs is acceptable on the login nodes. (Mostly sleeps; The actual jobs run on the compute nodes)pip installofvllmorflash-attnfrom source code on the login nodes is unacceptable (and is likely to fail anyways). (Takes far too much RAM to compile the CUDA kernels)Editing code with
nano,vimoremacsis acceptable. (Editors mostly sleep awaiting user keystrokes)Copying/moving files with
cp,mv, … is acceptable. (Mostly moves data)Connecting to compute nodes with
sshis acceptable. (Mostly sleeps, forwarding keystrokes and ports to/from the node)Using
tmuxis acceptable. (Mostly sleeps, managing the processes under its control)
Note
In a similar vein, you should not run VSCode remote SSH instances directly on login nodes, because even though they are typically not very computationally expensive, when many people do it, they add up! See Visual Studio Code for specific instructions.
SSH Private Keys
A private SSH key commonly takes the form of an obscure text file. It encodes the digital secret of how to make an electronic signature — specifically, yours. The content of a private SSH key might resemble
-----BEGIN OPENSSH PRIVATE KEY-----
b3BlbnNzaC1rZXktdjEAAAAABG5vbmUAAAAEbm9uZQAAAAAAAAABAAABlwAAAAdzc2gtcn
NhAAAAAwEAAQAAAYEAl5dD/UU2CvauaVS2/4/iWoUyO1Hey+m8KojCFMvIywL6PPdYRqVa
FOidmOw/E9V2HVzHz/z/2Dj6TO5xNX1qJFk7A/ACGGc1+KguIDQWdjR6AZb5Tat+aAMYro
…
aSeJOS59knbQJeBwPm0g5G+iFz6R17446dXk5jn3/29AutF5MPnKwqE0mjywxCLYxVX3He
YSOCZfE80P/z4sImW82BYxAzKtI8kKagLmHS4gXJEmE13Dfyq0xcB3q5OMuQ2fZwvukTx3
xdWgyqFrMyC4wHAAAAAAEC
-----END OPENSSH PRIVATE KEY-----
In the real world, a handwritten signature is useless for authenticating you if it can be easily reproduced by others. In the virtual world, the same is true. Anyone who has your private key is capable of reproducing your electronic signature. It is therefore essential that only one person — you — holds this private key. The secrecy of the private key is the guarantor of your online identity.
Mila will *never* ask you for your private SSH key, and any pretense of request for a private key constitutes an attempt at phishing and identity theft. Keep your private keys safe and do not share them with anyone. Do not put them in the cloud, your emails, Slack messages, or Git repos. Protect them with a passphrase.
SSH Public Keys
A public SSH key is a simple line of text, albeit sometimes very long, commonly
found in a file with the .pub extension. It encodes the digital knowledge
required to recognize and validate your electronic signature, without however
making it possible to reproduce it elsewhere. Here are three examples of public
SSH keys:
ssh-rsa AAAAB3NzaC1yc2EAAAADAQABAAABgQDMYpSndal/…mPL+NXs=
ssh-ed25519 AAAA…d/ca2h user@server
ecdsa-sha2-nistp256 AAAA…hWQcQg8= mylaptop
You are requested to submit just such a public SSH key to Mila, which will allow Mila to recognize you when you connect to the Mila cluster, but without revealing the secret of how to perform your signature.
Checking If You Already Have SSH (Private) Keys
Usually, a private SSH key is found in the hidden directory ~/.ssh/ and is
named id_rsa, id_ed25519, or id_ecdsa. Its corresponding public SSH
key is usually in the same directory and shares the same filename, except with
a .pub suffix.
Generating an SSH Private Key
If no private SSH key already exists, you can create one with the ssh-keygen utility:
RSA |
Ed25519 |
|---|---|
Integer factorization |
Elliptic curve |
|
|
|
|
Tip
The pass-phrase protects the SSH private key on-disk. The
passphrase is not the same thing as the pass-word used to log into
your personal computer account. However, choosing them to be equal may
allow for automatic unlocking of encrypted SSH private keys at login, in
combination with software such as pam_ssh(8) (Linux) or Keychain
(Mac OS X/macOS). This makes the good practice of using encrypted keys
convenient as well.
Generating an SSH Public Key from a Private Key
If a private SSH key exists, but not its corresponding SSH public key, it can
be recalculated with the ssh-keygen utility as well:
RSA |
Ed25519 |
|---|---|
SSH public key: >380 bytes @ 2048 bits (not rec.) >550 bytes @ 3072 bits (recommended) >725 bytes @ 4096 bits (slower) >1400 bytes @ 8192 bits (much slower) |
SSH public key: ~82 bytes |
|
|
It is this SSH public key that you should submit in the IT Onboarding Training form.
Configuring SSH
SSH uses a configuration file ~/.ssh/config (right next to the SSH keys)
to indicate which connection settings to use for each SSH server one can
connect to.
The Mila login nodes require:
Hostname:login.server.mila.quebecPort:2222User: Your Mila account usernamePreferredAuthentications:publickey,keyboard-interactive
Password authentication will be withdrawn on March 31, 2025.
A simple SSH configuration is automatically created and added for you to
~/.ssh/config by mila init.
Alternatively, more advanced users can edit the SSH .config file
manually.
Manual SSH configuration
If you would like to set entries in your ~/.ssh/config file manually for
advanced use-cases, you may use the following as inspiration:
# Mila
Host mila login.server.mila.quebec
Hostname login.server.mila.quebec
Host mila1 login-1.login.server.mila.quebec
Hostname login-1.login.server.mila.quebec
Host mila2 login-2.login.server.mila.quebec
Hostname login-2.login.server.mila.quebec
Host mila3 login-3.login.server.mila.quebec
Hostname login-3.login.server.mila.quebec
Host mila4 login-4.login.server.mila.quebec
Hostname login-4.login.server.mila.quebec
Host mila5 login-5.login.server.mila.quebec
Hostname login-5.login.server.mila.quebec
Host cn-????
Hostname %h.server.mila.quebec
Match host !*login.server.mila.quebec,*.server.mila.quebec
Hostname %h
ProxyJump mila
Match host *login.server.mila.quebec
Port 2222
ServerAliveInterval 120
ServerAliveCountMax 5
Match host *.server.mila.quebec
PreferredAuthentications publickey,keyboard-interactive
AddKeysToAgent yes
## Consider uncommenting:
# ForwardAgent yes
## Delete if on Linux, uncomment if on Mac:
# UseKeychain yes
User CHANGEME_YOUR_MILA_USERNAME
Important
Please make the required edits to the template above, especially regarding
CHANGEME_YOUR_MILA_USERNAME!
mila init
To make it easier to set up a productive environment, Mila publishes the
milatools package, which defines a mila init command which will
automatically perform some of the below steps for you. You can install it with
pip and use it, provided your Python version is at least 3.9:
pip install milatools
mila init
Note
This guide is current for milatools >= 0.0.17. If you have installed an older
version previously, run pip install -U milatools to upgrade and re-run
mila init in order to apply new features or bug fixes.
Connecting to compute nodes
If (and only if) you have a job running on compute node cnode, you are
allowed to SSH to it, if for some reason you need a second terminal.
That session will be automatically ended when your job ends.
First, however, you need to add your public key (the one you provided to IT-support)
to the ~/.ssh/authorized_keys file on the cluster, or configure an
ssh-agent that will forward that key when connecting to the compute node.
# ON A LOGIN NODE
mkdir -p ~/.ssh
echo "THE SSH PUBLIC KEY THAT YOU GAVE TO IT-SUPPORT" >> ~/.ssh/authorized_keys
chmod 600 ~/.ssh/authorized_keys
chmod 700 ~/.ssh
chmod go-w ~ # in case you accidentally gave too many permissions for $HOME in the past.
Then from the login node you can write ssh cnode. From your local
machine, you can use ssh -J mila USERNAME@cnode (-J represents a “jump”
through the login node, necessary because the compute nodes are behind a
firewall).
If you wish, you may also add the following wildcard rule in your .ssh/config:
Host *.server.mila.quebec !*login.server.mila.quebec
HostName %h
User YOUR-USERNAME
ProxyJump mila
This will let you connect to a compute node with ssh <node>.server.mila.quebec.
Auto-allocation with mila-cpu
If you install milatools and run mila init, then you can automatically allocate
a CPU on a compute node and connect to it by running:
ssh mila-cpu
And that’s it! Multiple connections to mila-cpu will all reuse the same job, so
you can use it liberally. It also works transparently with VSCode’s Remote SSH feature.
We recommend using this for light work that is too heavy for a login node but does not require a lot of resources: editing via VSCode, building conda environments, tests, etc.
The mila-cpu entry should be in your .ssh/config. Changes are at your own risk.
Using a non-Bash Unix shell
While Mila does not provide support in debugging your shell setup, Bash is the standard shell to be used on the cluster and the cluster is designed to support both Bash and Zsh shells. If you think things should work with Zsh and they don’t, please contact Mila’s IT support.
Running your code
SLURM commands guide
Basic Usage
The SLURM documentation provides extensive information on the available commands to query the cluster status or submit jobs.
Below are some basic examples of how to use SLURM.
Submitting jobs
Batch job
In order to submit a batch job, you have to create a script containing the main command(s) you would like to execute on the allocated resources/nodes.
1#!/bin/bash
2#SBATCH --job-name=test
3#SBATCH --output=job_output.txt
4#SBATCH --error=job_error.txt
5#SBATCH --ntasks=1
6#SBATCH --time=10:00
7#SBATCH --mem=100Gb
8
9module load python/3.5
10python my_script.py
Your job script is then submitted to SLURM with sbatch (ref.)
sbatch job_script
sbatch: Submitted batch job 4323674
The working directory of the job will be the one where your executed sbatch.
Tip
Slurm directives can be specified on the command line alongside sbatch or
inside the job script with a line starting with #SBATCH.
Interactive job
Workload managers usually run batch jobs to avoid having to watch its
progression and let the scheduler run it as soon as resources are available. If
you want to get access to a shell while leveraging cluster resources, you can
submit an interactive jobs where the main executable is a shell with the
srun/salloc (srun/salloc) commands
salloc
Will start an interactive job on the first node available with the default
resources set in SLURM (1 task/1 CPU). srun accepts the same arguments as
sbatch with the exception that the environment is not passed.
Tip
To pass your current environment to an interactive job, add
--preserve-env to srun.
salloc can also be used and is mostly a wrapper around srun if provided
without more info but it gives more flexibility if for example you want to get
an allocation on multiple nodes.
Job submission arguments
In order to accurately select the resources for your job, several arguments are available. The most important ones are:
Argument |
Description |
|---|---|
-n, –ntasks=<number> |
The number of task in your script, usually =1 |
-c, –cpus-per-task=<ncpus> |
The number of cores for each task |
-t, –time=<time> |
Time requested for your job |
–mem=<size[units]> |
Memory requested for all your tasks |
–gres=<list> |
Select generic resources such as GPUs for your job: |
Tip
Always consider requesting the adequate amount of resources to improve the scheduling of your job (small jobs always run first).
Checking job status
To display jobs currently in queue, use squeue and to get only your jobs type
squeue -u $USER
JOBID USER NAME ST START_TIME TIME NODES CPUS TRES_PER_NMIN_MEM NODELIST (REASON) COMMENT
133 my_username myjob R 2019-03-28T18:33 0:50 1 2 N/A 7000M node1 (None) (null)
Note
The maximum number of jobs able to be submitted to the system per user is 1000 (MaxSubmitJobs=1000) at any given time from the given association. If this limit is reached, new submission requests will be denied until existing jobs in this association complete.
Removing a job
To cancel your job simply use scancel
scancel 4323674
Partitioning
Since we don’t have many GPUs on the cluster, resources must be shared as fairly
as possible. The --partition=/-p flag of SLURM allows you to set the
priority you need for a job. Each job assigned with a priority can preempt jobs
with a lower priority: unkillable > main > long. Once preempted, your job is
killed without notice and is automatically re-queued on the same partition until
resources are available. (To leverage a different preemption mechanism, see the
Handling preemption)
Flag |
Max Resource Usage |
Max Time |
Note |
|---|---|---|---|
--partition=unkillable |
6 CPUs, mem=32G, 1 GPU |
2 days |
|
--partition=unkillable-cpu |
2 CPUs, mem=16G |
2 days |
CPU-only jobs |
--partition=short-unkillable |
mem=1000G, 4 GPUs |
3 hours (!) |
Large but short jobs |
--partition=main |
8 CPUs, mem=48G, 2 GPUs |
5 days |
|
--partition=main-cpu |
8 CPUs, mem=64G |
5 days |
CPU-only jobs |
--partition=long |
no limit of resources |
7 days |
|
--partition=long-cpu |
no limit of resources |
7 days |
CPU-only jobs |
Warning
Historically, before the 2022 introduction of CPU-only nodes (e.g. the cn-f
series), CPU jobs ran side-by-side with the GPU jobs on GPU nodes. To prevent
them obstructing any GPU job, they were always lowest-priority and preemptible.
This was implemented by automatically assigning them to one of the now-obsolete
partitions cpu_jobs, cpu_jobs_low or cpu_jobs_low-grace.
Do not use these partition names anymore. Prefer the *-cpu partition
names defined above.
For backwards-compatibility purposes, the legacy partition names are translated
to their effective equivalent long-cpu, but they will eventually be removed
entirely.
Note
As a convenience, should you request the unkillable, main or long
partition for a CPU-only job, the partition will be translated to its -cpu
equivalent automatically.
For instance, to request an unkillable job with 1 GPU, 4 CPUs, 10G of RAM and 12h of computation do:
sbatch --gres=gpu:1 -c 4 --mem=10G -t 12:00:00 --partition=unkillable <job.sh>
You can also make it an interactive job using salloc:
salloc --gres=gpu:1 -c 4 --mem=10G -t 12:00:00 --partition=unkillable
The Mila cluster has many different types of nodes/GPUs. To request a specific type of node/GPU, you can add specific feature requirements to your job submission command.
To access those special nodes you need to request them explicitly by adding the
flag --constraint=<name>. The full list of nodes in the Mila Cluster can be
accessed Node profile description.
Examples:
To request a machine with 2 GPUs using NVLink, you can use
sbatch -c 4 --gres=gpu:2 --constraint=nvlink
To request a DGX system with 8 A100 GPUs, you can use
sbatch -c 16 --gres=gpu:8 --constraint="dgx&ere"
Feature |
Particularities |
|---|---|
12gb/32gb/40gb/48gb/80gb |
Request a specific amount of GPU memory |
volta/turing/ampere |
Request a specific GPU architecture |
nvlink |
Machine with GPUs using the NVLink interconnect technology |
dgx |
NVIDIA DGX system with DGX OS |
Information on partitions/nodes
sinfo (ref.) provides most of the
information about available nodes and partitions/queues to submit jobs to.
Partitions are a group of nodes usually sharing similar features. On a partition, some job limits can be applied which will override those asked for a job (i.e. max time, max CPUs, etc…)
To display available partitions, simply use
sinfo
PARTITION AVAIL TIMELIMIT NODES STATE NODELIST
batch up infinite 2 alloc node[1,3,5-9]
batch up infinite 6 idle node[10-15]
cpu up infinite 6 idle cpu_node[1-15]
gpu up infinite 6 idle gpu_node[1-15]
To display available nodes and their status, you can use
sinfo -N -l
NODELIST NODES PARTITION STATE CPUS MEMORY TMP_DISK WEIGHT FEATURES REASON
node[1,3,5-9] 2 batch allocated 2 246 16000 0 (null) (null)
node[2,4] 2 batch drain 2 246 16000 0 (null) (null)
node[10-15] 6 batch idle 2 246 16000 0 (null) (null)
...
And to get statistics on a job running or terminated, use sacct with some of
the fields you want to display
sacct --format=User,JobID,Jobname,partition,state,time,start,end,elapsed,nnodes,ncpus,nodelist,workdir -u $USER
User JobID JobName Partition State Timelimit Start End Elapsed NNodes NCPUS NodeList WorkDir
--------- ------------ ---------- ---------- ---------- ---------- ------------------- ------------------- ---------- -------- ---------- --------------- --------------------
my_usern+ 2398 run_extra+ batch RUNNING 130-05:00+ 2019-03-27T18:33:43 Unknown 1-01:07:54 1 16 node9 /home/mila/my_usern+
my_usern+ 2399 run_extra+ batch RUNNING 130-05:00+ 2019-03-26T08:51:38 Unknown 2-10:49:59 1 16 node9 /home/mila/my_usern+
Or to get the list of all your previous jobs, use the --start=YYYY-MM-DD flag. You can check sacct(1) for further information about additional time formats.
sacct -u $USER --start=2019-01-01
scontrol (ref.) can be used to
provide specific information on a job (currently running or recently terminated)
scontrol show job 43123
JobId=43123 JobName=python_script.py
UserId=my_username(1500000111) GroupId=student(1500000000) MCS_label=N/A
Priority=645895 Nice=0 Account=my_username QOS=normal
JobState=RUNNING Reason=None Dependency=(null)
Requeue=1 Restarts=3 BatchFlag=1 Reboot=0 ExitCode=0:0
RunTime=2-10:41:57 TimeLimit=130-05:00:00 TimeMin=N/A
SubmitTime=2019-03-26T08:47:17 EligibleTime=2019-03-26T08:49:18
AccrueTime=2019-03-26T08:49:18
StartTime=2019-03-26T08:51:38 EndTime=2019-08-03T13:51:38 Deadline=N/A
PreemptTime=None SuspendTime=None SecsPreSuspend=0
LastSchedEval=2019-03-26T08:49:18
Partition=slurm_partition AllocNode:Sid=login-node-1:14586
ReqNodeList=(null) ExcNodeList=(null)
NodeList=node2
BatchHost=node2
NumNodes=1 NumCPUs=16 NumTasks=1 CPUs/Task=16 ReqB:S:C:T=0:0:*:*
TRES=cpu=16,mem=32000M,node=1,billing=3
Socks/Node=* NtasksPerN:B:S:C=1:0:*:* CoreSpec=*
MinCPUsNode=16 MinMemoryNode=32000M MinTmpDiskNode=0
Features=(null) DelayBoot=00:00:00
OverSubscribe=OK Contiguous=0 Licenses=(null) Network=(null)
WorkDir=/home/mila/my_username
StdErr=/home/mila/my_username/slurm-43123.out
StdIn=/dev/null
StdOut=/home/mila/my_username/slurm-43123.out
Power=
Or more info on a node and its resources
scontrol show node node9
NodeName=node9 Arch=x86_64 CoresPerSocket=4
CPUAlloc=16 CPUTot=16 CPULoad=1.38
AvailableFeatures=(null)
ActiveFeatures=(null)
Gres=(null)
NodeAddr=10.252.232.4 NodeHostName=mila20684000000 Port=0 Version=18.08
OS=Linux 4.15.0-1036 #38-Ubuntu SMP Fri Dec 7 02:47:47 UTC 2018
RealMemory=32000 AllocMem=32000 FreeMem=23262 Sockets=2 Boards=1
State=ALLOCATED+CLOUD ThreadsPerCore=2 TmpDisk=0 Weight=1 Owner=N/A MCS_label=N/A
Partitions=slurm_partition
BootTime=2019-03-26T08:50:01 SlurmdStartTime=2019-03-26T08:51:15
CfgTRES=cpu=16,mem=32000M,billing=3
AllocTRES=cpu=16,mem=32000M
CapWatts=n/a
CurrentWatts=0 LowestJoules=0 ConsumedJoules=0
ExtSensorsJoules=n/s ExtSensorsWatts=0 ExtSensorsTemp=n/s
Useful Commands
- salloc
Get an interactive job and give you a shell. (ssh like) CPU only
- salloc --gres=gpu:1 -c 2 --mem=12000
Get an interactive job with one GPU, 2 CPUs and 12000 MB RAM
- sbatch
start a batch job (same options as salloc)
- sattach --pty <jobid>.0
Re-attach a dropped interactive job
- sinfo
status of all nodes
- sinfo -Ogres:27,nodelist,features -tidle,mix,alloc
List GPU type and FEATURES that you can request
- savail
(Custom) List available gpu
- scancel <jobid>
Cancel a job
- squeue
summary status of all active jobs
- squeue -u $USER
summary status of all YOUR active jobs
- squeue -j <jobid>
summary status of a specific job
- squeue -Ojobid,name,username,partition,state,timeused,nodelist,gres,tres
status of all jobs including requested resources (see the SLURM squeue doc for all output options)
- scontrol show job <jobid>
Detailed status of a running job
- sacct -j <job_id> -o NodeList
Get the node where a finished job ran
- sacct -u $USER -S <start_time> -E <stop_time>
Find info about old jobs
- sacct -oJobID,JobName,User,Partition,Node,State
List of current and recent jobs
Special GPU requirements
Specific GPU architecture and memory can be easily requested through the
--gres flag by using either
--gres=gpu:architecture:number--gres=gpu:memory:number--gres=gpu:model:number
Example:
To request 1 GPU with at least 48GB of memory use
sbatch -c 4 --gres=gpu:48gb:1
The full list of GPU and their features can be accessed here.
Example script
Here is a sbatch script that follows good practices on the Mila cluster:
1#!/bin/bash
2
3#SBATCH --partition=unkillable # Ask for unkillable job
4#SBATCH --cpus-per-task=2 # Ask for 2 CPUs
5#SBATCH --gres=gpu:1 # Ask for 1 GPU
6#SBATCH --mem=10G # Ask for 10 GB of RAM
7#SBATCH --time=3:00:00 # The job will run for 3 hours
8#SBATCH -o /network/scratch/<u>/<username>/slurm-%j.out # Write the log on scratch
9
10# 1. Load the required modules
11module --quiet load anaconda/3
12
13# 2. Load your environment
14conda activate "<env_name>"
15
16# 3. Copy your dataset on the compute node
17cp /network/datasets/<dataset> $SLURM_TMPDIR
18
19# 4. Launch your job, tell it to save the model in $SLURM_TMPDIR
20# and look for the dataset into $SLURM_TMPDIR
21python main.py --path $SLURM_TMPDIR --data_path $SLURM_TMPDIR
22
23# 5. Copy whatever you want to save on $SCRATCH
24cp $SLURM_TMPDIR/<to_save> /network/scratch/<u>/<username>/
Portability concerns and solutions
When working on a software project, it is important to be aware of all the software and libraries the project relies on and to list them explicitly and under a version control system in such a way that they can easily be installed and made available on different systems. The upsides are significant:
Easily install and run on the cluster
Ease of collaboration
Better reproducibility
To achieve this, try to always keep in mind the following aspects:
Versions: For each dependency, make sure you have some record of the specific version you are using during development. That way, in the future, you will be able to reproduce the original environment which you know to be compatible. Indeed, the more time passes, the more likely it is that newer versions of some dependency have breaking changes. The
pip freezecommand can create such a record for Python dependencies.Isolation: Ideally, each of your software projects should be isolated from the others. What this means is that updating the environment for project A should not update the environment for project B. That way, you can freely install and upgrade software and libraries for the former without worrying about breaking the latter (which you might not notice until weeks later, the next time you work on project B!) Isolation can be made easy using UV, as well as Python Virtual environments and, as a last resort, Containers.
Managing your environments
Virtual environments
A virtual environment in Python is a local, isolated environment in which you can install or uninstall Python packages without interfering with the global environment (or other virtual environments). It usually lives in a directory (location varies depending on whether you use venv, conda or poetry). In order to use a virtual environment, you have to activate it. Activating an environment essentially sets environment variables in your shell so that:
pythonpoints to the right Python version for that environment (different virtual environments can use different versions of Python!)pythonlooks for packages in the virtual environmentpip installinstalls packages into the virtual environmentAny shell commands installed via
pip installare made available
To run experiments within a virtual environment, you can simply activate it
in the script given to sbatch.
Pip/Virtualenv
Pip is the most widely used package manager for Python and each cluster provides several Python versions through the associated module which comes with pip. In order to install new packages, you will first have to create a personal space for them to be stored. The usual solution (as it is the recommended solution on Digital Research Alliance of Canada clusters) is to use virtual environments, although UV is now the recommended way to manage Python installations, virtual environments and dependencies.
Note
We recommend you use UV to manage your Python virtual environments instead of doing it manually. The next section will give an overview of how to install it and use it.
First, load the Python module you want to use:
module load python/3.8
Then, create a virtual environment in your home directory:
python -m venv $HOME/<env>
Where <env> is the name of your environment. Finally, activate the environment:
source $HOME/<env>/bin/activate
You can now install any Python package you wish using the pip command, e.g.
pytorch:
pip install torch torchvision
Or Tensorflow:
pip install tensorflow-gpu
UV
In many cases, where your dependencies are Python packages, we highly recommend using UV, a modern package manager for Python.
In addition to all the same features as pip, it also manages Python installations, virtual environments, and makes your environments easier to reproduce and reuse across compute clusters.
Note
UV is not currently available as a module on the Mila or DRAC clusters at the time of writing. To use it, you first need to install it using this command on a cluster login node:
curl -LsSf https://astral.sh/uv/install.sh | sh
Pip/virtualenv command |
UV pip equivalent |
UV project command (recommended) |
|
|---|---|---|---|
Create your virtualenv |
|
||
Activate the virtualenv |
|
(same) |
(same, but often unnecessary) |
Install a package |
activate venv then |
||
Run a command
(ex. |
|
|
|
Where are dependencies declared? |
Maybe in a |
Maybe in a |
|
Easy to change Python versions? |
No |
somewhat |
Yes: |
While you can use UV as a drop-in replacement for pip, we recommend adopting a project-based workflow:
Use uv init to create a new project. A
pyproject.tomlfile will be created. This is where your dependencies are listed.uv init --python=3.12Use uv add to add (and uv remove to remove) dependencies to your project. This will update the
pyproject.tomlfile and update the virtual environment.uv add torch- Use uv run to run commands, for example
uv run python train.py. This will automatically do the following: Create or update the virtualenv (with the correct Python version) if necessary, based the dependencies in
pyproject.toml.Activates the virtualenv.
Runs the command you provided, e.g.
python train.py.
uv run python main.py
- Use uv run to run commands, for example
Conda
Another solution for Python is to use miniconda or anaconda which are also available through the module
command: (the use of Conda is not recommended for Digital Research Alliance of
Canada clusters due to the availability of custom-built packages for pip)
module load miniconda/3
=== Module miniconda/3 loaded ===]
o enable conda environment functions, first use:
To create an environment (see here for details) using a specific Python version, you may write:
conda create -n <env> python=3.9
Where <env> is the name of your environment. You can now activate it by doing:
conda activate <env>
You are now ready to install any Python package you want in this environment. For instance, to install PyTorch, you can find the Conda command of any version you want on pytorch’s website, e.g:
conda install pytorch torchvision cudatoolkit=10.0 -c pytorch
If you make a lot of environments and install/uninstall a lot of packages, it can be good to periodically clean up Conda’s cache:
conda clean -it
Mamba
When installing new packages with conda install, conda uses a built-in
dependency solver for solving the dependency graph of all packages (and their
versions) requested such that package dependency conflicts are avoided.
In some cases, especially when there are many packages already installed in a conda environment, conda’s built-in dependency solver can struggle to solve the dependency graph, taking several to tens of minutes, and sometimes never solving. In these cases, it is recommended to try libmamba.
To install and set the libmamba solver, run the following commands:
# Install miniconda
# (you can not use the preinstalled anaconda/miniconda as installing libmamba
# requires ownership over the anaconda/miniconda install directory)
wget https://repo.anaconda.com/miniconda/Miniconda3-py310_22.11.1-1-Linux-x86_64.sh
bash Miniconda3-py310_22.11.1-1-Linux-x86_64.sh
# Install libmamba
conda install -n base conda-libmamba-solver
By default, conda uses the built-in solver when installing packages, even after
installing other solvers. To try libmamba once, add --solver=libmamba in
your `conda install` command. For example:
conda install tensorflow --solver=libmamba
You can set libmamba as the default solver by adding solver: libmamba
to your .condarc configuration file located under your $HOME directory.
You can create it if it doesn’t exist. You can also run:
conda config --set solver libmamba
Using Modules
A lot of software, such as Python and Conda, is already compiled and available on
the cluster through the module command and its sub-commands. In particular,
if you wish to use Python 3.7 you can simply do:
module load python/3.7
The module command
For a list of available modules, simply use:
module avail
-------------------------------------------------------------------------------------------------------------- Global Aliases ---------------------------------------------------------------------------------------------------------------
cuda/10.0 -> cudatoolkit/10.0 cuda/9.2 -> cudatoolkit/9.2 pytorch/1.4.1 -> python/3.7/cuda/10.2/cudnn/7.6/pytorch/1.4.1 tensorflow/1.15 -> python/3.7/tensorflow/1.15
cuda/10.1 -> cudatoolkit/10.1 mujoco-py -> python/3.7/mujoco-py/2.0 pytorch/1.5.0 -> python/3.7/cuda/10.2/cudnn/7.6/pytorch/1.5.0 tensorflow/2.2 -> python/3.7/tensorflow/2.2
cuda/10.2 -> cudatoolkit/10.2 mujoco-py/2.0 -> python/3.7/mujoco-py/2.0 pytorch/1.5.1 -> python/3.7/cuda/10.2/cudnn/7.6/pytorch/1.5.1
cuda/11.0 -> cudatoolkit/11.0 pytorch -> python/3.7/cuda/10.2/cudnn/7.6/pytorch/1.5.1 tensorflow -> python/3.7/tensorflow/2.2
cuda/9.0 -> cudatoolkit/9.0 pytorch/1.4.0 -> python/3.7/cuda/10.2/cudnn/7.6/pytorch/1.4.0 tensorflow-cpu/1.15 -> python/3.7/tensorflow/1.15
-------------------------------------------------------------------------------------------------- /cvmfs/config.mila.quebec/modules/Core ---------------------------------------------------------------------------------------------------
Mila (S,L) anaconda/3 (D) go/1.13.5 miniconda/2 mujoco/1.50 python/2.7 python/3.6 python/3.8 singularity/3.0.3 singularity/3.2.1 singularity/3.5.3 (D)
anaconda/2 go/1.12.4 go/1.14 (D) miniconda/3 (D) mujoco/2.0 (D) python/3.5 python/3.7 (D) singularity/2.6.1 singularity/3.1.1 singularity/3.4.2
------------------------------------------------------------------------------------------------ /cvmfs/config.mila.quebec/modules/Compiler -------------------------------------------------------------------------------------------------
python/3.7/mujoco-py/2.0
-------------------------------------------------------------------------------------------------- /cvmfs/config.mila.quebec/modules/Cuda ---------------------------------------------------------------------------------------------------
cuda/10.0/cudnn/7.3 cuda/10.0/nccl/2.4 cuda/10.1/nccl/2.4 cuda/11.0/nccl/2.7 cuda/9.0/nccl/2.4 cudatoolkit/9.0 cudatoolkit/10.1 cudnn/7.6/cuda/10.0/tensorrt/7.0
cuda/10.0/cudnn/7.5 cuda/10.1/cudnn/7.5 cuda/10.2/cudnn/7.6 cuda/9.0/cudnn/7.3 cuda/9.2/cudnn/7.6 cudatoolkit/9.2 cudatoolkit/10.2 cudnn/7.6/cuda/10.1/tensorrt/7.0
cuda/10.0/cudnn/7.6 (D) cuda/10.1/cudnn/7.6 (D) cuda/10.2/nccl/2.7 cuda/9.0/cudnn/7.5 (D) cuda/9.2/nccl/2.4 cudatoolkit/10.0 cudatoolkit/11.0 (D) cudnn/7.6/cuda/9.0/tensorrt/7.0
------------------------------------------------------------------------------------------------ /cvmfs/config.mila.quebec/modules/Pytorch --------------------------------------------------------------------------------------------------
python/3.7/cuda/10.1/cudnn/7.6/pytorch/1.4.1 python/3.7/cuda/10.1/cudnn/7.6/pytorch/1.5.1 (D) python/3.7/cuda/10.2/cudnn/7.6/pytorch/1.5.0
python/3.7/cuda/10.1/cudnn/7.6/pytorch/1.5.0 python/3.7/cuda/10.2/cudnn/7.6/pytorch/1.4.1 python/3.7/cuda/10.2/cudnn/7.6/pytorch/1.5.1 (D)
----------------------------------------------------------------------------------------------- /cvmfs/config.mila.quebec/modules/Tensorflow ------------------------------------------------------------------------------------------------
python/3.7/tensorflow/1.15 python/3.7/tensorflow/2.0 python/3.7/tensorflow/2.2 (D)
Modules can be loaded using the load command:
module load <module>
To search for a module or a software, use the command spider:
module spider search_term
E.g.: by default, python2 will refer to the os-shipped installation of python2.7 and python3 to python3.6.
If you want to use python3.7 you can type:
module load python3.7
Available Software
Modules are divided in 5 main sections:
Section |
Description |
|---|---|
Core |
Base interpreter and software (Python, go, etc…) |
Compiler |
Interpreter-dependent software (see the note below) |
Cuda |
Toolkits, cudnn and related libraries |
Pytorch/Tensorflow |
Pytorch/TF built with a specific Cuda/Cudnn version for Mila’s GPUs (see the related paragraph) |
Note
Modules which are nested (../../..) usually depend on other software/module loaded alongside the main module. No need to load the dependent software, the complex naming scheme allows an automatic detection of the dependent module(s):
i.e.: Loading cudnn/7.6/cuda/9.0/tensorrt/7.0 will load cudnn/7.6 and
cuda/9.0 alongside
python/3.X is a particular dependency which can be served through
python/3.X or anaconda/3 and is not automatically loaded to let the
user pick his favorite flavor.
Default package location
Python by default uses the user site package first and packages provided by
module last to not interfere with your installation. If you want to skip
packages installed in your site-packages folder (in your /home directory), you
have to start Python with the -s flag.
To check which package is loaded at import, you can print package.__file__
to get the full path of the package.
Example:
module load pytorch/1.5.0
python -c 'import torch;print(torch.__file__)'
home/mila/my_home/.local/lib/python3.7/site-packages/torch/__init__.py <== package from your own site-package
Now with the -s flag:
module load pytorch/1.5.0
python -s -c 'import torch;print(torch.__file__)'
cvmfs/ai.mila.quebec/apps/x86_64/debian/pytorch/python3.7-cuda10.1-cudnn7.6-v1.5.0/lib/python3.7/site-packages/torch/__init__.py'
On using containers
Another option for creating portable code is Using containers.
Containers are a popular approach at deploying applications by packaging a lot of the required dependencies together. The most popular tool for this is Docker, but Docker cannot be used on the Mila cluster (nor the other clusters from Digital Research Alliance of Canada).
One popular mechanism for containerisation on a computational cluster is called Singularity. This is the recommended approach for running containers on the Mila cluster. See section Singularity for more details.
Using containers
Podman containers are now available as tech preview on the Mila cluster without root privileges using podman.
Generally any command-line argument accepted by docker will work with podman. This means that you can mostly use the docker examples you find on the web by replacing docker with podman in the command line.
Note
Complete Podman Documentation: https://docs.podman.io/en/stable/
Using in SLURM
To use podman you can just use the podman command in either a batch script or an interactive job.
One difference in configuration is that for certain technical reasons all the storage for podman (images, containers, …) is on a job-specific location and will be lost after the job is complete or preempted. If you have data that must be preseved across jobs, you can mount a local folder inside the container, such as $SCRATCH or your home to save data.
$ podman run --mount type=bind,source=$SCRATCH/exp,destination=/data/exp bash touch /data/exp/file
$ ls $SCRATCH/exp
file
You can use multiple containers in a single job, but you have to be careful about the memory and CPU limits of the job.
Note
Due to the cluster environment you may see warning messages like
WARN[0000] "/" is not a shared mount, this could cause issues or missing mounts with rootless containers,
ERRO[0000] cannot find UID/GID for user <user>: no subuid ranges found for user "<user>" in /etc/subuid - check rootless mode in man pages.,
WARN[0000] Using rootless single mapping into the namespace. This might break some images. Check /etc/subuid and /etc/subgid for adding sub*ids if not using a network user
or
WARN[0005] Failed to add pause process to systemd sandbox cgroup: dbus: couldn't determine address of session bus
but as far as we can see those can be safely ignored and should not have
an impact on your images.
GPU
To use a GPU in a container, you need a GPU job and then use --device
nvidia.com/gpu=all to make all GPUs allocated available in the container or
--device nvidia.com/gpu=N where N is the gpu index you want in the
container, starting at 0.
$ nvidia-smi
Fri Dec 13 12:47:34 2024
+-----------------------------------------------------------------------------------------+
| NVIDIA-SMI 560.35.03 Driver Version: 560.35.03 CUDA Version: 12.6 |
|-----------------------------------------+------------------------+----------------------+
| GPU Name Persistence-M | Bus-Id Disp.A | Volatile Uncorr. ECC |
| Fan Temp Perf Pwr:Usage/Cap | Memory-Usage | GPU-Util Compute M. |
| | | MIG M. |
|=========================================+========================+======================|
| 0 NVIDIA L40S On | 00000000:4A:00.0 Off | 0 |
| N/A 25C P8 36W / 350W | 1MiB / 46068MiB | 0% Default |
| | | N/A |
+-----------------------------------------+------------------------+----------------------+
| 1 NVIDIA L40S On | 00000000:61:00.0 Off | 0 |
| N/A 26C P8 35W / 350W | 1MiB / 46068MiB | 0% Default |
| | | N/A |
+-----------------------------------------+------------------------+----------------------+
+-----------------------------------------------------------------------------------------+
| Processes: |
| GPU GI CI PID Type Process name GPU Memory |
| ID ID Usage |
|=========================================================================================|
| No running processes found |
+-----------------------------------------------------------------------------------------+
$ podman run --device nvidia.com/gpu=all nvidia/cuda:11.6.1-base-ubuntu20.04 nvidia-smi
Fri Dec 13 17:48:21 2024
+-----------------------------------------------------------------------------------------+
| NVIDIA-SMI 560.35.03 Driver Version: 560.35.03 CUDA Version: 12.6 |
|-----------------------------------------+------------------------+----------------------+
| GPU Name Persistence-M | Bus-Id Disp.A | Volatile Uncorr. ECC |
| Fan Temp Perf Pwr:Usage/Cap | Memory-Usage | GPU-Util Compute M. |
| | | MIG M. |
|=========================================+========================+======================|
| 0 NVIDIA L40S On | 00000000:4A:00.0 Off | 0 |
| N/A 25C P8 36W / 350W | 1MiB / 46068MiB | 0% Default |
| | | N/A |
+-----------------------------------------+------------------------+----------------------+
| 1 NVIDIA L40S On | 00000000:61:00.0 Off | 0 |
| N/A 25C P8 35W / 350W | 1MiB / 46068MiB | 0% Default |
| | | N/A |
+-----------------------------------------+------------------------+----------------------+
+-----------------------------------------------------------------------------------------+
| Processes: |
| GPU GI CI PID Type Process name GPU Memory |
| ID ID Usage |
|=========================================================================================|
| No running processes found |
+-----------------------------------------------------------------------------------------+
$ podman run --device nvidia.com/gpu=0 nvidia/cuda:11.6.1-base-ubuntu20.04 nvidia-smi
Fri Dec 13 17:48:33 2024
+-----------------------------------------------------------------------------------------+
| NVIDIA-SMI 560.35.03 Driver Version: 560.35.03 CUDA Version: 12.6 |
|-----------------------------------------+------------------------+----------------------+
| GPU Name Persistence-M | Bus-Id Disp.A | Volatile Uncorr. ECC |
| Fan Temp Perf Pwr:Usage/Cap | Memory-Usage | GPU-Util Compute M. |
| | | MIG M. |
|=========================================+========================+======================|
| 0 NVIDIA L40S On | 00000000:4A:00.0 Off | 0 |
| N/A 25C P8 36W / 350W | 1MiB / 46068MiB | 0% Default |
| | | N/A |
+-----------------------------------------+------------------------+----------------------+
+-----------------------------------------------------------------------------------------+
| Processes: |
| GPU GI CI PID Type Process name GPU Memory |
| ID ID Usage |
|=========================================================================================|
| No running processes found |
+-----------------------------------------------------------------------------------------+
$ podman run --device nvidia.com/gpu=1 nvidia/cuda:11.6.1-base-ubuntu20.04 nvidia-smi
Fri Dec 13 17:48:40 2024
+-----------------------------------------------------------------------------------------+
| NVIDIA-SMI 560.35.03 Driver Version: 560.35.03 CUDA Version: 12.6 |
|-----------------------------------------+------------------------+----------------------+
| GPU Name Persistence-M | Bus-Id Disp.A | Volatile Uncorr. ECC |
| Fan Temp Perf Pwr:Usage/Cap | Memory-Usage | GPU-Util Compute M. |
| | | MIG M. |
|=========================================+========================+======================|
| 0 NVIDIA L40S On | 00000000:61:00.0 Off | 0 |
| N/A 25C P8 35W / 350W | 1MiB / 46068MiB | 0% Default |
| | | N/A |
+-----------------------------------------+------------------------+----------------------+
+-----------------------------------------------------------------------------------------+
| Processes: |
| GPU GI CI PID Type Process name GPU Memory |
| ID ID Usage |
|=========================================================================================|
| No running processes found |
+-----------------------------------------------------------------------------------------+
You can pass --device multiple times to add more than one gpus to the container.
Note
CDI (GPU) support documentation: https://docs.nvidia.com/datacenter/cloud-native/container-toolkit/latest/cdi-support.html#running-a-workload-with-cdi
Singularity
Overview
What is Singularity?
Running Docker on SLURM is a security problem (e.g. running as root, being able to mount any directory). The alternative is to use Singularity, which is a popular solution in the world of HPC.
There is a good level of compatibility between Docker and Singularity, and we can find many exaggerated claims about able to convert containers from Docker to Singularity without any friction. Oftentimes, Docker images from DockerHub are 100% compatible with Singularity, and they can indeed be used without friction, but things get messy when we try to convert our own Docker build files to Singularity recipes.
Links to official documentation
official Singularity user guide (this is the one you will use most often)
official Singularity admin guide
Overview of the steps used in practice
Most often, the process to create and use a Singularity container is:
on your Linux computer (at home or work)
select a Docker image from DockerHub (e.g. pytorch/pytorch)
make a recipe file for Singularity that starts with that DockerHub image
build the recipe file, thus creating the image file (e.g.
my-pytorch-image.sif)test your singularity container before send it over to the cluster
rsync -av my-pytorch-image.sif <login-node>:Documents/my-singularity-images
on the login node for that cluster
queue your jobs with
sbatch ...(note that your jobs will copy over the
my-pytorch-image.sifto $SLURM_TMPDIR and will then launch Singularity with that image)do something else while you wait for them to finish
queue more jobs with the same
my-pytorch-image.sif, reusing it many times over
In the following sections you will find specific examples or tips to accomplish in practice the steps highlighted above.
Nope, not on MacOS
Singularity does not work on MacOS, as of the time of this writing in 2021. Docker does not actually run on MacOS, but there Docker silently installs a virtual machine running Linux, which makes it a pleasant experience, and the user does not need to care about the details of how Docker does it.
Given its origins in HPC, Singularity does not provide that kind of seamless experience on MacOS, even though it’s technically possible to run it inside a Linux virtual machine on MacOS.
Where to build images
Building Singularity images is a rather heavy task, which can take 20 minutes if you have a lot of steps in your recipe. This makes it a bad task to run on the login nodes of our clusters, especially if it needs to be run regularly.
On the Mila cluster, we are lucky to have unrestricted internet access on the compute nodes, which means that anyone can request an interactive CPU node (no need for GPU) and build their images there without problem.
Warning
Do not build Singularity images from scratch every time your run a
job in a large batch. This will be a colossal waste of GPU time as well as
internet bandwidth. If you setup your workflow properly (e.g. using bind
paths for your code and data), you can spend months reusing the same
Singularity image my-pytorch-image.sif.
Building the containers
Building a container is like creating a new environment except that containers are much more powerful since they are self-contained systems. With singularity, there are two ways to build containers.
The first one is by yourself, it’s like when you got a new Linux laptop and you don’t really know what you need, if you see that something is missing, you install it. Here you can get a vanilla container with Ubuntu called a sandbox, you log in and you install each packages by yourself. This procedure can take time but will allow you to understand how things work and what you need. This is recommended if you need to figure out how things will be compiled or if you want to install packages on the fly. We’ll refer to this procedure as singularity sandboxes.
The second way is more like you know what you want, so you write a list of everything you need, you send it to singularity and it will install everything for you. Those lists are called singularity recipes.
First way: Build and use a sandbox
You might ask yourself: On which machine should I build a container?
First of all, you need to choose where you’ll build your container. This operation requires memory and high cpu usage.
Warning
Do NOT build containers on any login nodes !
(Recommended for beginner) If you need to use apt-get, you should build the container on your laptop with sudo privileges. You’ll only need to install singularity on your laptop. Windows/Mac users can look there and Ubuntu/Debian users can use directly:
sudo apt-get install singularity-containerIf you can’t install singularity on your laptop and you don’t need apt-get, you can reserve a cpu node on the Mila cluster to build your container.
In this case, in order to avoid too much I/O over the network, you should define the singularity cache locally:
export SINGULARITY_CACHEDIR=$SLURM_TMPDIR
If you can’t install singularity on your laptop and you want to use apt-get, you can use singularity-hub to build your containers and read Recipe_section.
Download containers from the web
Hopefully, you may not need to create containers from scratch as many have been already built for the most common deep learning software. You can find most of them on dockerhub.
Go on dockerhub and select the container you want to pull.
For example, if you want to get the latest PyTorch version with GPU support (Replace runtime by devel if you need the full Cuda toolkit):
singularity pull docker://pytorch/pytorch:1.0.1-cuda10.0-cudnn7-runtime
Or the latest TensorFlow:
singularity pull docker://tensorflow/tensorflow:latest-gpu-py3
Currently the pulled image pytorch.simg or tensorflow.simg is read-only
meaning that you won’t be able to install anything on it. Starting now, PyTorch
will be taken as example. If you use TensorFlow, simply replace every
pytorch occurrences by tensorflow.
How to add or install stuff in a container
The first step is to transform your read only container
pytorch-1.0.1-cuda10.0-cudnn7-runtime.simg in a writable version that will
allow you to add packages.
Warning
Depending on the version of singularity you are using, singularity will build a container with the extension .simg or .sif. If you’re using .sif files, replace every occurences of .simg by .sif.
Tip
If you want to use apt-get you have to put sudo ahead of the following commands
This command will create a writable image in the folder pytorch.
singularity build --sandbox pytorch pytorch-1.0.1-cuda10.0-cudnn7-runtime.simg
Then you’ll need the following command to log inside the container.
singularity shell --writable -H $HOME:/home pytorch
Once you get into the container, you can use pip and install anything you need
(Or with apt-get if you built the container with sudo).
Warning
Singularity mounts your home folder, so if you install things into
the $HOME of your container, they will be installed in your real
$HOME!
You should install your stuff in /usr/local instead.
Creating useful directories
One of the benefits of containers is that you’ll be able to use them across different clusters. However for each cluster the datasets and experiments folder location can be different. In order to be invariant to those locations, we will create some useful mount points inside the container:
mkdir /dataset
mkdir /tmp_log
mkdir /final_log
From now, you won’t need to worry anymore when you write your code to specify
where to pick up your dataset. Your dataset will always be in /dataset
independently of the cluster you are using.
Testing
If you have some code that you want to test before finalizing your container, you have two choices. You can either log into your container and run Python code inside it with:
singularity shell --nv pytorch
Or you can execute your command directly with
singularity exec --nv pytorch Python YOUR_CODE.py
Tip
—nv allows the container to use gpus. You don’t need this if you don’t plan to use a gpu.
Warning
Don’t forget to clear the cache of the packages you installed in the containers.
Creating a new image from the sandbox
Once everything you need is installed inside the container, you need to convert it back to a read-only singularity image with:
singularity build pytorch_final.simg pytorch
Second way: Use recipes
A singularity recipe is a file including specifics about installation software, environment variables, files to add, and container metadata. It is a starting point for designing any custom container. Instead of pulling a container and installing your packages manually, you can specify in this file the packages you want and then build your container from this file.
Here is a toy example of a singularity recipe installing some stuff:
################# Header: Define the base system you want to use ################
# Reference of the kind of base you want to use (e.g., docker, debootstrap, shub).
Bootstrap: docker
# Select the docker image you want to use (Here we choose tensorflow)
From: tensorflow/tensorflow:latest-gpu-py3
################# Section: Defining the system #################################
# Commands in the %post section are executed within the container.
%post
echo "Installing Tools with apt-get"
apt-get update
apt-get install -y cmake libcupti-dev libyaml-dev wget unzip
apt-get clean
echo "Installing things with pip"
pip install tqdm
echo "Creating mount points"
mkdir /dataset
mkdir /tmp_log
mkdir /final_log
# Environment variables that should be sourced at runtime.
%environment
# use bash as default shell
SHELL=/bin/bash
export SHELL
A recipe file contains two parts: the header and sections. In the
header you specify which base system you want to use, it can be any docker
or singularity container. In sections, you can list the things you want to
install in the subsection post or list the environment’s variable you need
to source at each runtime in the subsection environment. For a more detailed
description, please look at the singularity documentation.
In order to build a singularity container from a singularity recipe file, you should use:
sudo singularity build <NAME_CONTAINER> <YOUR_RECIPE_FILES>
Warning
You always need to use sudo when you build a container from a recipe. As there is no access to sudo on the cluster, a personal computer or the use singularity hub is needed to build a container
Build recipe on singularity hub
Singularity hub allows users to build containers from recipes directly on singularity-hub’s cloud meaning that you don’t need to build containers by yourself. You need to register on singularity-hub and link your singularity-hub account to your GitHub account, then:
Create a new github repository.
Add a collection on singularity-hub and select the github repository your created.
Clone the github repository on your computer.
$ git clone <url>Write the singularity recipe and save it as a file named Singularity.
Git add Singularity, commit and push on the master branch
$ git add Singularity $ git commit $ git push origin master
At this point, robots from singularity-hub will build the container for you, you will be able to download your container from the website or directly with:
singularity pull shub://<github_username>/<repository_name>
Example: Recipe with OpenAI gym, MuJoCo and Miniworld
Here is an example on how you can use a singularity recipe to install complex environment such as OpenAI gym, MuJoCo and Miniworld on a PyTorch based container. In order to use MuJoCo, you’ll need to copy the key stored on the Mila cluster in /ai/apps/mujoco/license/mjkey.txt to your current directory.
#This is a dockerfile that sets up a full Gym install with test dependencies
Bootstrap: docker
# Here we ll build our container upon the pytorch container
From: pytorch/pytorch:1.0-cuda10.0-cudnn7-runtime
# Now we'll copy the mjkey file located in the current directory inside the container's root
# directory
%files
mjkey.txt
# Then we put everything we need to install
%post
export PATH=$PATH:/opt/conda/bin
apt -y update && \
apt install -y keyboard-configuration && \
apt install -y \
python3-dev \
python-pyglet \
python3-opengl \
libhdf5-dev \
libjpeg-dev \
libboost-all-dev \
libsdl2-dev \
libosmesa6-dev \
patchelf \
ffmpeg \
xvfb \
libhdf5-dev \
openjdk-8-jdk \
wget \
git \
unzip && \
apt clean && \
rm -rf /var/lib/apt/lists/*
pip install h5py
# Download Gym and MuJoCo
mkdir /Gym && cd /Gym
git clone https://github.com/openai/gym.git || true && \
mkdir /Gym/.mujoco && cd /Gym/.mujoco
wget https://www.roboti.us/download/mjpro150_linux.zip && \
unzip mjpro150_linux.zip && \
wget https://www.roboti.us/download/mujoco200_linux.zip && \
unzip mujoco200_linux.zip && \
mv mujoco200_linux mujoco200
# Export global environment variables
export MUJOCO_PY_MJKEY_PATH=/Gym/.mujoco/mjkey.txt
export MUJOCO_PY_MUJOCO_PATH=/Gym/.mujoco/mujoco150/
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/Gym/.mujoco/mjpro150/bin
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/Gym/.mujoco/mujoco200/bin
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/usr/local/bin
cp /mjkey.txt /Gym/.mujoco/mjkey.txt
# Install Python dependencies
wget https://raw.githubusercontent.com/openai/mujoco-py/master/requirements.txt
pip install -r requirements.txt
# Install Gym and MuJoCo
cd /Gym/gym
pip install -e '.[all]'
# Change permission to use mujoco_py as non sudoer user
chmod -R 777 /opt/conda/lib/python3.6/site-packages/mujoco_py/
pip install --upgrade minerl
# Export global environment variables
%environment
export SHELL=/bin/sh
export MUJOCO_PY_MJKEY_PATH=/Gym/.mujoco/mjkey.txt
export MUJOCO_PY_MUJOCO_PATH=/Gym/.mujoco/mujoco150/
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/Gym/.mujoco/mjpro150/bin
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/Gym/.mujoco/mujoco200/bin
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/usr/local/bin
export PATH=/Gym/gym/.tox/py3/bin:$PATH
%runscript
exec /bin/sh "$@"
Here is the same recipe but written for TensorFlow:
#This is a dockerfile that sets up a full Gym install with test dependencies
Bootstrap: docker
# Here we ll build our container upon the tensorflow container
From: tensorflow/tensorflow:latest-gpu-py3
# Now we'll copy the mjkey file located in the current directory inside the container's root
# directory
%files
mjkey.txt
# Then we put everything we need to install
%post
apt -y update && \
apt install -y keyboard-configuration && \
apt install -y \
python3-setuptools \
python3-dev \
python-pyglet \
python3-opengl \
libjpeg-dev \
libboost-all-dev \
libsdl2-dev \
libosmesa6-dev \
patchelf \
ffmpeg \
xvfb \
wget \
git \
unzip && \
apt clean && \
rm -rf /var/lib/apt/lists/*
# Download Gym and MuJoCo
mkdir /Gym && cd /Gym
git clone https://github.com/openai/gym.git || true && \
mkdir /Gym/.mujoco && cd /Gym/.mujoco
wget https://www.roboti.us/download/mjpro150_linux.zip && \
unzip mjpro150_linux.zip && \
wget https://www.roboti.us/download/mujoco200_linux.zip && \
unzip mujoco200_linux.zip && \
mv mujoco200_linux mujoco200
# Export global environment variables
export MUJOCO_PY_MJKEY_PATH=/Gym/.mujoco/mjkey.txt
export MUJOCO_PY_MUJOCO_PATH=/Gym/.mujoco/mujoco150/
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/Gym/.mujoco/mjpro150/bin
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/Gym/.mujoco/mujoco200/bin
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/usr/local/bin
cp /mjkey.txt /Gym/.mujoco/mjkey.txt
# Install Python dependencies
wget https://raw.githubusercontent.com/openai/mujoco-py/master/requirements.txt
pip install -r requirements.txt
# Install Gym and MuJoCo
cd /Gym/gym
pip install -e '.[all]'
# Change permission to use mujoco_py as non sudoer user
chmod -R 777 /usr/local/lib/python3.5/dist-packages/mujoco_py/
# Then install miniworld
cd /usr/local/
git clone https://github.com/maximecb/gym-miniworld.git
cd gym-miniworld
pip install -e .
# Export global environment variables
%environment
export SHELL=/bin/bash
export MUJOCO_PY_MJKEY_PATH=/Gym/.mujoco/mjkey.txt
export MUJOCO_PY_MUJOCO_PATH=/Gym/.mujoco/mujoco150/
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/Gym/.mujoco/mjpro150/bin
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/Gym/.mujoco/mujoco200/bin
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/usr/local/bin
export PATH=/Gym/gym/.tox/py3/bin:$PATH
%runscript
exec /bin/bash "$@"
Keep in mind that those environment variables are sourced at runtime and not at
build time. This is why, you should also define them in the %post section
since they are required to install MuJoCo.
Using containers on clusters
How to use containers on clusters
On every cluster with Slurm, datasets and intermediate results should go in
$SLURM_TMPDIR while the final experiment results should go in $SCRATCH.
In order to use the container you built, you need to copy it on the cluster you
want to use.
Warning
You should always store your container in $SCRATCH !
Then reserve a node with srun/sbatch, copy the container and your dataset on the
node given by SLURM (i.e in $SLURM_TMPDIR) and execute the code
<YOUR_CODE> within the container <YOUR_CONTAINER> with:
singularity exec --nv -H $HOME:/home -B $SLURM_TMPDIR:/dataset/ -B $SLURM_TMPDIR:/tmp_log/ -B $SCRATCH:/final_log/ $SLURM_TMPDIR/<YOUR_CONTAINER> python <YOUR_CODE>
Remember that /dataset, /tmp_log and /final_log were created in the
previous section. Now each time, we’ll use singularity, we are explicitly
telling it to mount $SLURM_TMPDIR on the cluster’s node in the folder
/dataset inside the container with the option -B such that each dataset
downloaded by PyTorch in /dataset will be available in $SLURM_TMPDIR.
This will allow us to have code and scripts that are invariant to the cluster
environment. The option -H specify what will be the container’s home. For
example, if you have your code in $HOME/Project12345/Version35/ you can
specify -H $HOME/Project12345/Version35:/home, thus the container will only
have access to the code inside Version35.
If you want to run multiple commands inside the container you can use:
singularity exec --nv -H $HOME:/home -B $SLURM_TMPDIR:/dataset/ \
-B $SLURM_TMPDIR:/tmp_log/ -B $SCRATCH:/final_log/ \
$SLURM_TMPDIR/<YOUR_CONTAINER> bash -c 'pwd && ls && python <YOUR_CODE>'
Example: Interactive case (srun/salloc)
Once you get an interactive session with SLURM, copy <YOUR_CONTAINER> and
<YOUR_DATASET> to $SLURM_TMPDIR
0. Get an interactive session
srun --gres=gpu:1
1. Copy your container on the compute node
rsync -avz $SCRATCH/<YOUR_CONTAINER> $SLURM_TMPDIR
2. Copy your dataset on the compute node
rsync -avz $SCRATCH/<YOUR_DATASET> $SLURM_TMPDIR
Then use singularity shell to get a shell inside the container
3. Get a shell in your environment
singularity shell --nv \
-H $HOME:/home \
-B $SLURM_TMPDIR:/dataset/ \
-B $SLURM_TMPDIR:/tmp_log/ \
-B $SCRATCH:/final_log/ \
$SLURM_TMPDIR/<YOUR_CONTAINER>
4. Execute your code
python <YOUR_CODE>
or use singularity exec to execute <YOUR_CODE>.
3. Execute your code
singularity exec --nv \
-H $HOME:/home \
-B $SLURM_TMPDIR:/dataset/ \
-B $SLURM_TMPDIR:/tmp_log/ \
-B $SCRATCH:/final_log/ \
$SLURM_TMPDIR/<YOUR_CONTAINER> \
python <YOUR_CODE>
You can create also the following alias to make your life easier.
alias my_env='singularity exec --nv \
-H $HOME:/home \
-B $SLURM_TMPDIR:/dataset/ \
-B $SLURM_TMPDIR:/tmp_log/ \
-B $SCRATCH:/final_log/ \
$SLURM_TMPDIR/<YOUR_CONTAINER>'
This will allow you to run any code with:
my_env python <YOUR_CODE>
Example: sbatch case
You can also create a sbatch script:
:linenos:
#!/bin/bash
#SBATCH --cpus-per-task=6 # Ask for 6 CPUs
#SBATCH --gres=gpu:1 # Ask for 1 GPU
#SBATCH --mem=10G # Ask for 10 GB of RAM
#SBATCH --time=0:10:00 # The job will run for 10 minutes
# 1. Copy your container on the compute node
rsync -avz $SCRATCH/<YOUR_CONTAINER> $SLURM_TMPDIR
# 2. Copy your dataset on the compute node
rsync -avz $SCRATCH/<YOUR_DATASET> $SLURM_TMPDIR
# 3. Executing your code with singularity
singularity exec --nv \
-H $HOME:/home \
-B $SLURM_TMPDIR:/dataset/ \
-B $SLURM_TMPDIR:/tmp_log/ \
-B $SCRATCH:/final_log/ \
$SLURM_TMPDIR/<YOUR_CONTAINER> \
python "<YOUR_CODE>"
# 4. Copy whatever you want to save on $SCRATCH
rsync -avz $SLURM_TMPDIR/<to_save> $SCRATCH
Issue with PyBullet and OpenGL libraries
If you are running certain gym environments that require pyglet, you may
encounter a problem when running your singularity instance with the Nvidia
drivers using the --nv flag. This happens because the --nv flag also
provides the OpenGL libraries:
libGL.so.1 => /.singularity.d/libs/libGL.so.1
libGLX.so.0 => /.singularity.d/libs/libGLX.so.0
If you don’t experience those problems with pyglet, you probably don’t need
to address this. Otherwise, you can resolve those problems by apt-get install
-y libosmesa6-dev mesa-utils mesa-utils-extra libgl1-mesa-glx, and then making
sure that your LD_LIBRARY_PATH points to those libraries before the ones in
/.singularity.d/libs.
%environment
# ...
export LD_LIBRARY_PATH=/usr/lib/x86_64-linux-gnu/mesa:$LD_LIBRARY_PATH
Mila cluster
On the Mila cluster $SCRATCH is not yet defined, you should add the
experiment results you want to keep in /network/scratch/<u>/<username>/. In
order to use the sbatch script above and to match other cluster environment’s
names, you can define $SCRATCH as an alias for
/network/scratch/<u>/<username> with:
echo "export SCRATCH=/network/scratch/${USER:0:1}/$USER" >> ~/.bashrc
Then, you can follow the general procedure explained above.
Digital Research Alliance of Canada
Using singularity on Digital Research Alliance of Canada is similar except that
you need to add Yoshua’s account name and load singularity. Here is an example
of a sbatch script using singularity on compute Canada cluster:
Warning
You should use singularity/2.6 or singularity/3.4. There is a bug in singularity/3.2 which makes gpu unusable.
1#!/bin/bash
2#SBATCH --account=rpp-bengioy # Yoshua pays for your job
3#SBATCH --cpus-per-task=6 # Ask for 6 CPUs
4#SBATCH --gres=gpu:1 # Ask for 1 GPU
5#SBATCH --mem=32G # Ask for 32 GB of RAM
6#SBATCH --time=0:10:00 # The job will run for 10 minutes
7#SBATCH --output="/scratch/<user>/slurm-%j.out" # Modify the output of sbatch
8
9# 1. You have to load singularity
10module load singularity
11# 2. Then you copy the container to the local disk
12rsync -avz $SCRATCH/<YOUR_CONTAINER> $SLURM_TMPDIR
13# 3. Copy your dataset on the compute node
14rsync -avz $SCRATCH/<YOUR_DATASET> $SLURM_TMPDIR
15# 4. Executing your code with singularity
16singularity exec --nv \
17 -H $HOME:/home \
18 -B $SLURM_TMPDIR:/dataset/ \
19 -B $SLURM_TMPDIR:/tmp_log/ \
20 -B $SCRATCH:/final_log/ \
21 $SLURM_TMPDIR/<YOUR_CONTAINER> \
22 python "<YOUR_CODE>"
23# 5. Copy whatever you want to save on $SCRATCH
24rsync -avz $SLURM_TMPDIR/<to_save> $SCRATCH
Contributing datasets
If a dataset could help the research of others at Mila, this form can be filled to request its addition to /network/datasets.
Data Transmission using Globus Connect Personal
Mila doesn’t own a Globus license but if the source or destination provides a Globus account, like Digital Research Alliance of Canada for example, it’s possible to setup Globus Connect Personal to create a personal endpoint on the Mila cluster by following the Globus guide to Install, Configure, and Uninstall Globus Connect Personal for Linux.
This endpoint can then be used to transfer data to and from the Mila cluster.
JupyterHub
JupyterHub is a platform connected to SLURM to start a JupyterLab session as a batch job then connects it when the allocation has been granted. It does not require any ssh tunnel or port redirection, the hub acts as a proxy server that will redirect you to a session as soon as it is available.
It is currently available for Mila clusters and some Digital Research Alliance of Canada (Alliance) clusters.
Cluster |
Address |
Login type |
|---|---|---|
Mila Local |
Google Oauth |
|
Alliance |
DRAC login |
Warning
Do not forget to close the JupyterLab session! Closing the window leaves running the session and the SLURM job it is linked to.
To close it, use the hub menu and then Control Panel > Stop my server
Note
For Mila Clusters:
mila.quebec account credentials should be used to login and start a JupyterLab session.
Access Mila Storage in JupyterLab
Unfortunately, JupyterLab does not allow the navigation to parent directories of
$HOME. This makes some file systems like /network/datasets or
$SLURM_TMPDIR unavailable through their absolute path in the interface. It
is however possible to create symbolic links to those resources. To do so, you
can use the ln -s command:
ln -s /network/datasets $HOME
Note that $SLURM_TMPDIR is a directory that is dynamically created for each
job so you would need to recreate the symbolic link every time you start a
JupyterHub session:
ln -sf $SLURM_TMPDIR $HOME
Advanced SLURM usage and Multiple GPU jobs
Handling preemption
On the Mila cluster, jobs can preempt one-another depending on their priority (unkillable>high>low) (See the Slurm documentation)
The default preemption mechanism is to kill and re-queue the job automatically
without any notice. To allow a different preemption mechanism, every partition
have been duplicated (i.e. have the same characteristics as their counterparts)
allowing a 120sec grace period before killing your job but don’t requeue
it automatically: those partitions are referred by the suffix: -grace
(main-grace, long-grace, main-cpu-grace, long-cpu-grace).
When using a partition with a grace period, a series of signals consisting of
first SIGCONT and SIGTERM then SIGKILL will be sent to the SLURM
job. It’s good practice to catch those signals using the Linux trap command
to properly terminate a job and save what’s necessary to restart the job. On
each cluster, you’ll be allowed a grace period before SLURM actually kills
your job (SIGKILL).
The easiest way to handle preemption is by trapping the SIGTERM signal
1#SBATCH --ntasks=1
2#SBATCH ....
3
4exit_script() {
5 echo "Preemption signal, saving myself"
6 trap - SIGTERM # clear the trap
7 # Optional: sends SIGTERM to child/sub processes
8 kill -- -$$
9}
10
11trap exit_script SIGTERM
12
13# The main script part
14python3 my_script
Note
sbatch command insideexit_script function.Packing jobs
Multiple Nodes
Data Parallel
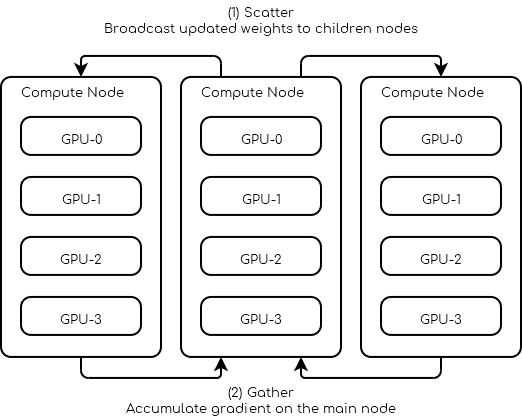
Request 3 nodes with at least 4 GPUs each.
1#!/bin/bash
2
3# Number of Nodes
4#SBATCH --nodes=3
5
6# Number of tasks. 3 (1 per node)
7#SBATCH --ntasks=3
8
9# Number of GPU per node
10#SBATCH --gres=gpu:4
11#SBATCH --gpus-per-node=4
12
13# 16 CPUs per node
14#SBATCH --cpus-per-gpu=4
15
16# 16Go per nodes (4Go per GPU)
17#SBATCH --mem=16G
18
19# we need all nodes to be ready at the same time
20#SBATCH --wait-all-nodes=1
21
22# Total resources:
23# CPU: 16 * 3 = 48
24# RAM: 16 * 3 = 48 Go
25# GPU: 4 * 3 = 12
26
27# Setup our rendez-vous point
28RDV_ADDR=$(hostname)
29WORLD_SIZE=$SLURM_JOB_NUM_NODES
30# -----
31
32srun -l torchrun \
33 --nproc_per_node=$SLURM_GPUS_PER_NODE\
34 --nnodes=$WORLD_SIZE\
35 --rdzv_id=$SLURM_JOB_ID\
36 --rdzv_backend=c10d\
37 --rdzv_endpoint=$RDV_ADDR\
38 training_script.py
You can find below a pytorch script outline on what a multi-node trainer could look like.
import os
import torch.distributed as dist
class Trainer:
def __init__(self):
self.local_rank = None
self.chk_path = ...
self.model = ...
@property
def device_id(self):
return self.local_rank
def load_checkpoint(self, path):
self.chk_path = path
# ...
def should_checkpoint(self):
# Note: only one worker saves its weights
return self.global_rank == 0 and self.local_rank == 0
def save_checkpoint(self):
if self.chk_path is None:
return
# Save your states here
# Note: you should save the weights of self.model not ddp_model
# ...
def initialize(self):
self.global_rank = int(os.environ.get("RANK", -1))
self.local_rank = int(os.environ.get("LOCAL_RANK", -1))
assert self.global_rank >= 0, 'Global rank should be set (Only Rank 0 can save checkpoints)'
assert self.local_rank >= 0, 'Local rank should be set'
dist.init_process_group(backend="gloo|nccl")
def sync_weights(self, resuming=False):
if resuming:
# in the case of resuming all workers need to load the same checkpoint
self.load_checkpoint()
# Wait for everybody to finish loading the checkpoint
dist.barrier()
return
# Make sure all workers have the same initial weights
# This makes the leader save his weights
if self.should_checkpoint():
self.save_checkpoint()
# All workers wait for the leader to finish
dist.barrier()
# All followers load the leader's weights
if not self.should_checkpoint():
self.load_checkpoint()
# Leader waits for the follower to load the weights
dist.barrier()
def dataloader(self, dataset, batch_size):
train_sampler = ElasticDistributedSampler(dataset)
train_loader = DataLoader(
dataset,
batch_size=batch_size,
num_workers=4,
pin_memory=True,
sampler=train_sampler,
)
return train_loader
def train_step(self):
# Your batch processing step here
# ...
pass
def train(self, dataset, batch_size):
self.sync_weights()
ddp_model = torch.nn.parallel.DistributedDataParallel(
self.model,
device_ids=[self.device_id],
output_device=self.device_id
)
loader = self.dataloader(dataset, batch_size)
for epoch in range(100):
for batch in iter(loader):
self.train_step(batch)
if self.should_checkpoint():
self.save_checkpoint()
def main():
trainer = Trainer()
trainer.load_checkpoint(path)
tainer.initialize()
trainer.train(dataset, batch_size)
Note
To bypass Python GIL (Global interpreter lock) pytorch spawn one process for each GPU. In the example above this means at least 12 processes are spawn, at least 4 on each node.
Weight and Biases (WandB)
Students supervised by core professors are elligible to the Mila organization on wandb. To request access, write to it-support@mila.quebec. Then please follow the guidelines below to get your account created or linked to Mila’s organization.
Logging in for the first time
For those who already have a WandB account
Note
If you already have an account and want to have access to mila-org with it, first add your email @mila.quebec to your account and make it your primary email. See documentation here on how to do so. Then log out so that you can make your first connection with single sign-on. Make sure to do this before following the next steps otherwise you will end up with 2 separate accounts
Go to https://wandb.ai and click sign in.
Enter your email @mila.quebec at the bottom.
The password box should disappear when you are done typing your email. Then click log in, and you will be redirected to a single sign-on page.
Select your account mila.quebec. If you already have an account, wandb will offer to link it, otherwise, you will be invited to create a new account.
For new accounts, select Professional.
Comet
Students supervised by core professors are elligible to the Mila organization on Comet. To request access, write to it-support@mila.quebec. Then please follow the guidelines below to get your account created within Mila’s organization. This account will be independant from your personal account if you already have one.
Logging in for the first time
To access mila-org, you need to login using the url https://comet.mila.quebec/. On first login, one of the following will apply:
If you have no Comet account or another account with an email address other than @mila.quebec: Comet will create a new account.
If you have an account with our email address @mila.quebec. Comet will link your account to mila-org.
Frequently asked questions (FAQs)
Connection/SSH issues
I’m getting connection refused while trying to connect to a login node
Login nodes are protected against brute force attacks and might ban your IP if it detects too many connections/failures. You will be automatically unbanned after 1 hour. For any further problem, please submit a support ticket.
Shell issues
How do I change my shell ?
By default you will be assigned /bin/bash as a shell. If you would like to
change for another one, please submit a support ticket.
SLURM issues
How can I get an interactive shell on the cluster ?
Use salloc [--slurm_options] without any executable at the end of the
command, this will launch your default shell on an interactive session. Remember
that an interactive session is bound to the login node where you start it so you
could risk losing your job if the login node becomes unreachable.
How can I reset my cluster password ?
To reset your password, please submit a support ticket.
Warning: your cluster password is the same as your Google Workspace account. So, after reset, you must use the new password for all your Google services.
srun: error: –mem and –mem-per-cpu are mutually exclusive
You can safely ignore this, salloc has a default memory flag in case you
don’t provide one.
How can I see where and if my jobs are running ?
Use squeue -u YOUR_USERNAME to see all your job status and locations.
To get more info on a running job, try scontrol show job #JOBID
Unable to allocate resources: Invalid account or account/partition combination specified
Chances are your account is not setup properly. You should submit a support ticket.
How do I cancel a job?
To cancel a specific job, use
scancel #JOBIDTo cancel all your jobs (running and pending), use
scancel -u YOUR_USERNAMETo cancel all your pending jobs only, use
scancel -t PD
How can I access a node on which one of my jobs is running ?
You can ssh into a node on which you have a job running, your ssh connection
will be adopted by your job, i.e. if your job finishes your ssh connection will
be automatically terminated. In order to connect to a node, you need to have
password-less ssh either with a key present in your home or with an
ssh-agent. You can generate a key on the login node like this:
ssh-keygen (3xENTER)
cat ~/.ssh/id_rsa.pub >> ~/.ssh/authorized_keys
chmod 600 ~/.ssh/authorized_keys
chmod 700 ~/.ssh
The ECDSA, RSA and ED25519 fingerprints for Mila’s compute nodes are:
SHA256:hGH64v72h/c0SfngAWB8WSyMj8WSAf5um3lqVsa7Cfk (ECDSA)
SHA256:4Es56W5ANNMQza2sW2O056ifkl8QBvjjNjfMqpB7/1U (RSA)
SHA256:gUQJw6l1lKjM1cCyennetPoQ6ST0jMhQAs/57LhfakA (ED25519)
I’m getting Permission denied (publickey) while trying to connect to a node
See previous question
Where do I put my data during a job ?
Your /home as well as the datasets are on shared file-systems, it is
recommended to copy them to the $SLURM_TMPDIR to better process them and
leverage higher-speed local drives. If you run a low priority job subject to
preemption, it’s better to save any output you want to keep on the shared file
systems, because the $SLURM_TMPDIR is deleted at the end of each job.
slurmstepd: error: Detected 1 oom-kill event(s) in step #####.batch cgroup
You exceeded the amount of memory allocated to your job, either you did not
request enough memory or you have a memory leak in your process. Try increasing
the amount of memory requested with --mem= or --mem-per-cpu=.
PyTorch issues
I randomly get INTERNAL ASSERT FAILED at "../aten/src/ATen/MapAllocator.cpp":263
You are using PyTorch 1.10.x and hitting #67864,
for which the solution is PR #72232
merged in PyTorch 1.11.x. For an immediate fix, consider the following compilable Gist:
hack.cpp.
Compile the patch to hack.so and then export LD_PRELOAD=/absolute/path/to/hack.so
before executing the Python process that import torch a broken PyTorch 1.10.
For Hydra users who are using the submitit launcher plug-in, the env_set key cannot
be used to set LD_PRELOAD in the environment as it does so too late at runtime. The
dynamic loader reads LD_PRELOAD only once and very early during the startup of any
process, before the variable can be set from inside the process. The hack must therefore
be injected using the setup key in Hydra YAML config file:
hydra:
launcher:
setup:
- export LD_PRELOAD=/absolute/path/to/hack.so
On MIG GPUs, I get torch.cuda.device_count() == 0 despite torch.cuda.is_available()
You are using PyTorch 1.13.x and hitting #90543, for which the solution is PR #92315 merged in PyTorch 2.0.
To avoid thus problem, update to PyTorch 2.0. If PyTorch 1.13.x is required, a workaround is to add the following to your script:
unset CUDA_VISIBLE_DEVICES
But this is no longer necessary with PyTorch >= 2.0.
I am told my PyTorch job abuses the filesystem with extreme amounts of IOPS
A fairly common issue in PyTorch is:
RuntimeError: one of the variables needed for gradient computation has been
modified by an inplace operation: [torch.cuda.FloatTensor [1, 50, 300]],
which is output 0 of SplitBackward, is at version 2; expected version 0
instead. Hint: enable anomaly detection to find the operation that failed to
compute its gradient, with torch.autograd.set_detect_anomaly(True).
PyTorch’s autograd engine contains an “anomaly detection mode”, which detects such things as NaN/infinities being created, and helps debugging in-place Tensor modifications. It is activated with
torch.autograd.set_detect_anomaly(True)
PyTorch’s implementation of the anomaly-detection mode tracks where every Tensor was created in the program. This involves the collection of the backtrace at the point the Tensor was created.
Unfortunately, the collection of a backtrace involves a stat() system
call to every source file in the backtrace. This is considered a metadata
access to $HOME and results in intolerably heavy traffic to the shared
filesystem containing the source code, usually $HOME, whatever the location
of the dataset, and even if it is on $SLURM_TMPDIR. It is the source-code
files being polled, not the dataset. As there can be hundreds of PyTorch tensors
created per iteration and thousands of iterations per second, this mode results
in extreme amounts of IOPS to the filesystem.
Warning
Do not use
torch.autograd.set_detect_anomaly(True)except for debugging an individual job interactively, and switch it off as soon as done using it.Do not set
torch.autograd.set_detect_anomaly(True)enabled unconditionally in all your jobs. It is not a consequence-free aid. Due to heavy use of filesystem calls, it has a performance impact and slows down your code, on top of abusing the filesystem.You will be contacted if you violate these guidelines due to the severity of its impact on shared filesystems.
Conda refuses to create an environment with Your installed CUDA driver is: not available
Anaconda attempts to auto-detect the NVIDIA driver version of the system and thus the maximum CUDA toolkit supported, in an attempt at choosing an appropriate CUDA Toolkit version.
However, on login and CPU nodes, there is no NVIDIA GPU and thus no need for
NVIDIA drivers. But that means conda’s auto-detection will not work on
those nodes, and packages declaring a minimum requirement on the drivers will
fail to install.
The solution in such a situation is to set the environment variable
CONDA_OVERRIDE_CUDA to the desired CUDA Toolkit version; For example,
CONDA_OVERRIDE_CUDA=11.8 conda create -n ENVNAME python=3.10 pytorch torchvision torchaudio pytorch-cuda=11.8 -c pytorch -c nvidia
This and other CONDA_OVERRIDE_* variables are documented in the conda manual.